Enabling d3plot2hdf5 to export results from DEM analyses has been a recent feature request. The following model of a funnel injection is directly taken from the LS-DYNA examples homepage.
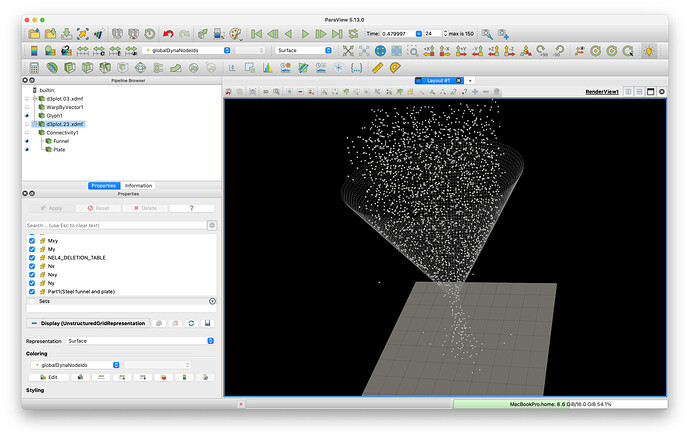
Below is a screenshot of the model in ParaView
The state file can be downloaded here: funnelInjection.pvsm (1.1 MB)
Once the simulation with LS-DYNA is finished, we end up with these d3plot database files:
ll -h d3plot*
-rw-r--r--@ 1 larski larski 2.6M Jul 8 2024 d3plot
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot01
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot02
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot03
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot04
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot05
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot06
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot07
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot08
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot09
-rw-r--r--@ 1 larski larski 133M Jul 8 2024 d3plot10
-rw-r--r--@ 1 larski larski 8.9M Jul 8 2024 d3plot11
-rw-r--r--@ 1 larski larski 8.9M Jul 8 2024 d3plot12
The converter is simply invoked by
d3plot2hdf5 d3plot
which then produces this output
+--------------------------------------------------------------------+
| |
| T A I L S I T |
| ------------- |
| |
| Program :: d3plot2hdf5 |
| Purpose :: convert d3plot to h5/xdmf |
| Build :: v0.10.0 [6fa1851] |
| Date :: Apr 28 2025,15:55:30 |
| News, discussions, bugs :: https://discourse.tailsit.com |
| |
| Terms: https://tailsit.com/downloads/TAILSIT_B2B_GTC.pdf |
| |
| For non-commercial use only |
| |
| Copyright, 2025 by TAILSIT GMBH. All rights reserved. |
| www.tailsit.com |
| |
+--------------------------------------------------------------------+
Options:
-- timeSteps: all
-- split files: false
Title/Part/Contact information:
-- Header: Funnel Injection
-- Parts:
[mat-id:1] Steel funnel and plate
[mat-id:2] DES
-- Contacts: (NONE)
LS-DYNA files:
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot01"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot02"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot03"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot04"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot05"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot06"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot07"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot08"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot09"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot10"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot11"
-- "/home/larski/git/d3plot2hdf5/tests/dem/FunnelInjection/complete/d3plot12"
LS-DYNA file information:
-- Title: Funnel Injection
-- Version: DEV
-- Creation date: Mon Jul 1 09:18:32 2024
-- Format: double precision
Additional ASCII Data:
-- NONE
State information:
-- max. no. of states found: 152
-- actual no. of states found: 152
-- state range: [0:151]
Output written to:
-- d3plot.03.xdmf
-- d3plot.23.xdmf
-- d3plot.h5
N O R M A L T E R M I N A T I O N 04/28/25 16:02:31
The converter took about 4 seconds to produce these files:
ll -h *.xdmf *.h5
-rw-rw-r-- 1 larski larski 1.4M Apr 26 07:38 d3plot.03.xdmf
-rw-rw-r-- 1 larski larski 1.3M Apr 26 07:38 d3plot.23.xdmf
-rw-rw-r-- 1 larski larski 1.5G Apr 26 07:38 d3plot.h5
All the heavy data is stored in the hdf5-file d3plot.h5 while the .xdmf files contain lightweight references to the .h5 file. The .xdmf files can then directly be processed by tools like ParaView.
The particle data is referenced from d3plot.03.xdmf while references to the structural data are stored in d3plot.23.xdmf.
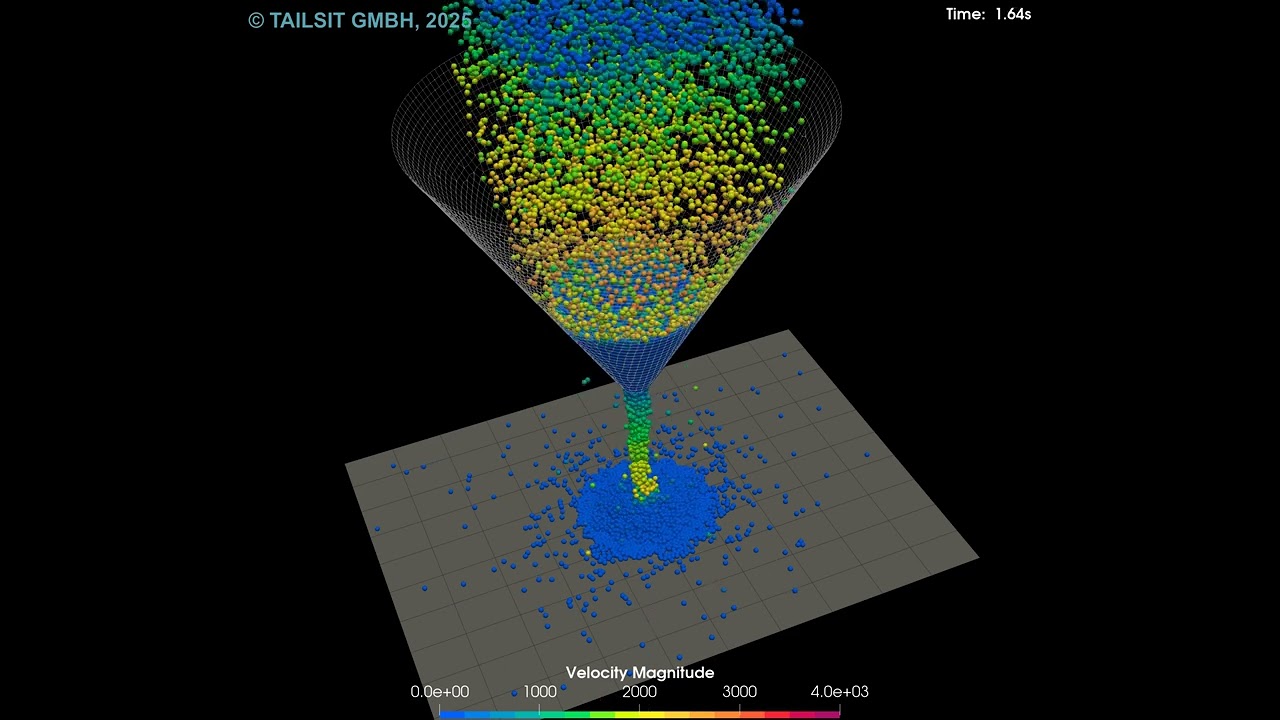
Below is a tiny animation of the results. It displays the particles’ velocities through a period of roughly 2.8 seconds.
Scripting
A word on scripting. .xdmf stores references to the data in such a way that ParaView can digest the heavy data. However, it is the .h5 file that stores the data iteself. So, if you want to script your post-processing tasks it is a good idea to get accustomed to the structure of the .h5 file.
We recommend using command line tools to investigate the contents of the .h5 file. On Debian based systems those command line tools can be installed with
sudo apt install hdf5-helpers
Other Linux distributions supply the hdf5-helpers, too. Simply use the respective package manager. Alternatively, those tools are typically bundled with the binary distributions of HDF5. Simply go to
for further information. This destination might be helpful for Windows installations, too.
Once the hdf5 tools are installed you can work, e.g., with h5dump or h5ls. The latter gives you a condensed view of the contents while the output of h5dump can be rather exhaustive. In many ways the HDF5 format can be viewed as a kind of filesystem, and this is exactly how h5ls behaves like.
So, let’s get started. Calling
h5ls d3plot.h5 | column
is likely to produce this lengthy output:
annotation Group d3plot_stp025 Group d3plot_stp051 Group d3plot_stp077 Group d3plot_stp103 Group d3plot_stp129 Group
d3plot_stp000 Group d3plot_stp026 Group d3plot_stp052 Group d3plot_stp078 Group d3plot_stp104 Group d3plot_stp130 Group
d3plot_stp001 Group d3plot_stp027 Group d3plot_stp053 Group d3plot_stp079 Group d3plot_stp105 Group d3plot_stp131 Group
d3plot_stp002 Group d3plot_stp028 Group d3plot_stp054 Group d3plot_stp080 Group d3plot_stp106 Group d3plot_stp132 Group
d3plot_stp003 Group d3plot_stp029 Group d3plot_stp055 Group d3plot_stp081 Group d3plot_stp107 Group d3plot_stp133 Group
d3plot_stp004 Group d3plot_stp030 Group d3plot_stp056 Group d3plot_stp082 Group d3plot_stp108 Group d3plot_stp134 Group
d3plot_stp005 Group d3plot_stp031 Group d3plot_stp057 Group d3plot_stp083 Group d3plot_stp109 Group d3plot_stp135 Group
d3plot_stp006 Group d3plot_stp032 Group d3plot_stp058 Group d3plot_stp084 Group d3plot_stp110 Group d3plot_stp136 Group
d3plot_stp007 Group d3plot_stp033 Group d3plot_stp059 Group d3plot_stp085 Group d3plot_stp111 Group d3plot_stp137 Group
d3plot_stp008 Group d3plot_stp034 Group d3plot_stp060 Group d3plot_stp086 Group d3plot_stp112 Group d3plot_stp138 Group
d3plot_stp009 Group d3plot_stp035 Group d3plot_stp061 Group d3plot_stp087 Group d3plot_stp113 Group d3plot_stp139 Group
d3plot_stp010 Group d3plot_stp036 Group d3plot_stp062 Group d3plot_stp088 Group d3plot_stp114 Group d3plot_stp140 Group
d3plot_stp011 Group d3plot_stp037 Group d3plot_stp063 Group d3plot_stp089 Group d3plot_stp115 Group d3plot_stp141 Group
d3plot_stp012 Group d3plot_stp038 Group d3plot_stp064 Group d3plot_stp090 Group d3plot_stp116 Group d3plot_stp142 Group
d3plot_stp013 Group d3plot_stp039 Group d3plot_stp065 Group d3plot_stp091 Group d3plot_stp117 Group d3plot_stp143 Group
d3plot_stp014 Group d3plot_stp040 Group d3plot_stp066 Group d3plot_stp092 Group d3plot_stp118 Group d3plot_stp144 Group
d3plot_stp015 Group d3plot_stp041 Group d3plot_stp067 Group d3plot_stp093 Group d3plot_stp119 Group d3plot_stp145 Group
d3plot_stp016 Group d3plot_stp042 Group d3plot_stp068 Group d3plot_stp094 Group d3plot_stp120 Group d3plot_stp146 Group
d3plot_stp017 Group d3plot_stp043 Group d3plot_stp069 Group d3plot_stp095 Group d3plot_stp121 Group d3plot_stp147 Group
d3plot_stp018 Group d3plot_stp044 Group d3plot_stp070 Group d3plot_stp096 Group d3plot_stp122 Group d3plot_stp148 Group
d3plot_stp019 Group d3plot_stp045 Group d3plot_stp071 Group d3plot_stp097 Group d3plot_stp123 Group d3plot_stp149 Group
d3plot_stp020 Group d3plot_stp046 Group d3plot_stp072 Group d3plot_stp098 Group d3plot_stp124 Group d3plot_stp150 Group
d3plot_stp021 Group d3plot_stp047 Group d3plot_stp073 Group d3plot_stp099 Group d3plot_stp125 Group d3plot_stp151 Group
d3plot_stp022 Group d3plot_stp048 Group d3plot_stp074 Group d3plot_stp100 Group d3plot_stp126 Group narbs Group
d3plot_stp023 Group d3plot_stp049 Group d3plot_stp075 Group d3plot_stp101 Group d3plot_stp127 Group
d3plot_stp024 Group d3plot_stp050 Group d3plot_stp076 Group d3plot_stp102 Group d3plot_stp128 Group
As you can see there are 152 time steps stored in d3plot.h5. If you now want to investigate a single time step a bit further you can do:
h5ls d3plot.h5/d3plot_stp000
which will give you:
fields03 Group
fields23 Group
grid03 Group
grid23 Group
gridXY stores the geometry information (or links to it), while fieldsXY stores the simulation data itself. Let’s investigate the latter a bit further. X denotes the reference dimension while Y denotes the dimension of the physical world. Hence, fields03 stores the particle data. Since this is the data we are interested in, we proceed with
h5ls d3plot.h5/d3plot_stp000/fields03
and obtain
cell Group
grid Group
node Group
These three groups denote to which entities the data is attached to. In a Finite Element simulation node would denote nodal or point data, cell would be element data, and grid would collect data such as energies or time series values which are global quantities that can neither be attached to a cell nor to a node.
Let’s assume we are further interested in the grid data. This time we invoke h5ls with the -r option which avoids further cycles and lists all groups recursively. So, entering
h5ls -r d3plot.h5/d3plot_stp000/fields03/grid
yields
/scalar Group
/scalar/internal\ energy(PART_0) Dataset {1}
/scalar/internal\ energy(PART_1) Dataset {1}
/scalar/internal\ energy(PART_ALL) Dataset {1}
/scalar/kinetic\ energy(PART_0) Dataset {1}
/scalar/kinetic\ energy(PART_1) Dataset {1}
/scalar/kinetic\ energy(PART_ALL) Dataset {1}
/scalar/mass(PART_0) Dataset {1}
/scalar/mass(PART_1) Dataset {1}
/scalar/time Dataset {1}
/scalar/total\ energy(PART_0) Dataset {1}
/scalar/total\ energy(PART_1) Dataset {1}
/scalar/total\ energy(PART_ALL) Dataset {1}
/vector Group
/vector/rigid\ body\ velocity(PART_0) Dataset {3}
/vector/rigid\ body\ velocity(PART_1) Dataset {3}
/vector/rigid\ body\ velocity(PART_ALL) Dataset {3}
This time we get to the data itself. We can see that grid contains a scalar and a vector group. So, if we are interested in the energies over time we now know exactly where the data of interest is stored. Clearly, you do not need to process the data soley with h5ls. You could also use software packages like, e.g., octave, Matlab, or Mathematica to accomplish this task. Feel free in the choice of your tools. And if you want to use a freely available tool that provides a graphical user interface we recommend HDFView.
Finally, this scripting section would be rather incomplete without a script itself. Now that we know the structure of the .h5 file we can create a simple Python-script that plots the energies over time. The script looks like:
#!/usr/bin/env python3
import h5py
import re
import matplotlib.pyplot as plt
import numpy as np
import sys
def extract_data( h5file: str, part: str ):
data = []
with h5py.File( h5file, 'r' ) as f:
group_names = list(f.keys())
pattern = re.compile( r'^d3plot_stp\d{3}$')
for group_name in group_names:
if pattern.match(group_name):
base = f"/{group_name}/fields03/grid/scalar"
# time
time = f"{base}/time"
# kinetic energy
ke = f"{base}/kinetic energy({part})"
# internal energy
ie = f"{base}/internal energy({part})"
# total energy
te = f"{base}/total energy({part})"
t = f[time][:][0]
kw = f[ke][:][0]
iw = f[ie][:][0]
tw = f[te][:][0]
data.append( [t, kw, iw, tw] )
return np.array(data)
def plot( data, part ):
t = data[:,0]
kw = data[:,1]
iw = data[:,2]
tw = data[:,3]
plt.figure(figsize=(12,8))
plt.plot(t,tw,marker='o', linestyle='-',color='b',label='Total')
plt.plot(t,iw, linestyle='-',color='g',label='Internal')
plt.plot(t,kw, linestyle='-',color='m',label='Kinetic')
plt.xlabel('time [s]')
plt.ylabel('energy')
plt.title(f'Energies over time [{part}]')
plt.grid(True)
plt.legend()
plt.savefig(f'energy_{part}.png', format='png', dpi=300)
plt.show()
if __name__ == "__main__":
try:
if len(sys.argv) is not 3:
raise ValueError(f"Usage: {sys.argv[0]} h5-file part, part = 0|1|ALL")
#
# PARSE COMMAND LINE
h5file = sys.argv[1]
part = "PART_"
part = f"{part}{sys.argv[2]}"
#
## EXTRACT DATA
data = extract_data( h5file, part )
#
# PLOT DATA
plot(data,part)
#
# DONE
except ValueError as e:
print( f"An error has ocurred: {e}")
The script can be downloaded here:
processEnergies.py (2.0 KB)
And the required packages can be found herein:
requirements.txt (255 Bytes)
Calling
python3 -m venv venv
source venv/bin/activate
pip3 install -r requirements.txt
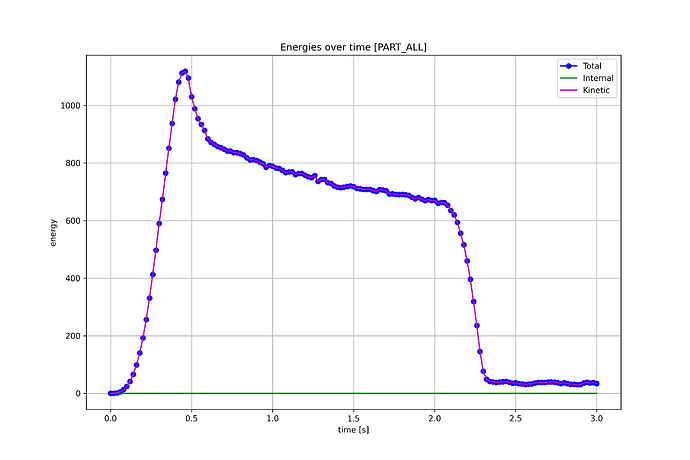
./processEnergies.py d3plot.h5 ALL
then produces the following plot
That’s it!